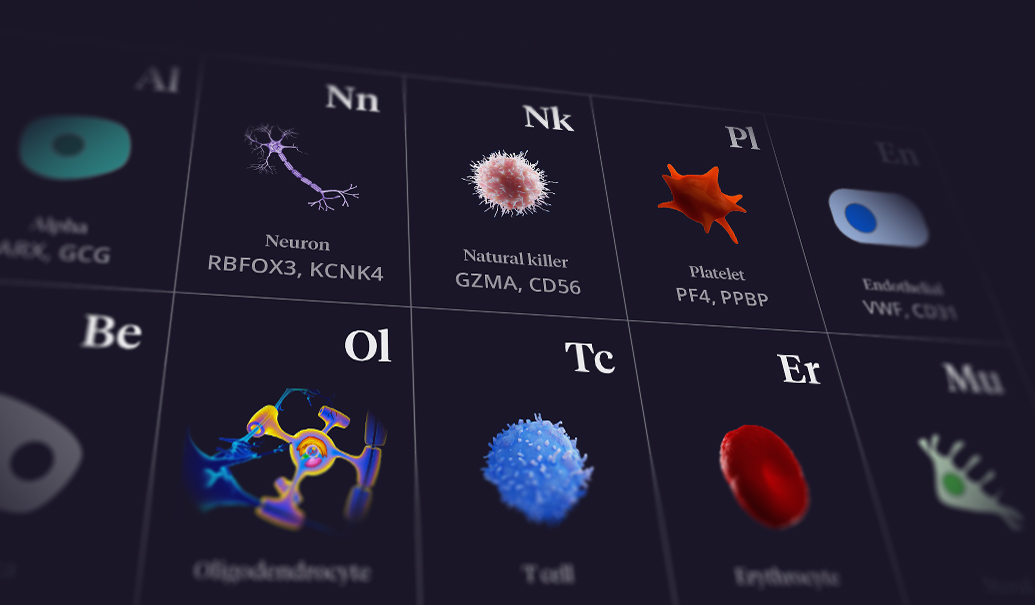
The demand for new ways to develop drugs has become a tired call to action. The pharmaceutical industry has recognized for years that low-hanging fruit in therapeutic targets no longer exist. Similarly, the concerningly large proportion of drugs that fail phase 2 clinical trials due to lack of efficacy is not a new challenge. Yet billions of dollars are spent each year on drugs that ultimately fail clinical trials. Why?
A growing proportion of the industry recognizes that these failures are not just due to challenges in clinical trial design, patient retention or ineffective animal models—all of which are significant challenges and are being addressed by technological and molecular advances. Yet the industry has not effectively addressed two big contributors to drug failure: the incomplete understanding of the molecular mechanisms behind disease and the inability to find the patient subpopulation for which a drug actually works.
Advancing drug development with high-resolution single-cell data
These two challenges remain because traditional approaches to studying disease and identifying therapeutic targets remain centered around bulk sequencing or AI and machine-learning-enabled in silico target identification efforts that either cannot or do not take into account the fact that genes and proteins do not act in isolation. “Many are still approaching drug development from a 10,000-foot view even though we now have the technology to explore at a microscopic level,” said Rasmus Wernersson, principal, executive director of bioinformatics and scientific engineering at ZS.
The ability to sequence DNA and RNA from single cells—one of the greatest scientific advances of the last decade—enables scientists to study cells at the finest resolution possible. This is the resolution required to identify the molecular mechanisms driving disease and to identify the specific types of cells for which a drug works.
While deeply exploring single-cell data, researchers at ZS helped identify a new type of cell sharing a common progenitor with adipogenic cells, the therapeutic implications of which are vast and include approaches like toggling between different cellular development states. We predict that the industry is about to experience a watershed moment that will change how new drugs are identified and developed, and this will drastically reduce the clinical trial failure rate that we have struggled with for so long.
Single-cell data is the key to completing biological maps
Since being named “2013 Method of the Year” by Nature, single-cell DNA and RNA sequencing have become routine. Where once sequencing the tiny amount of genetic material contained within a single cell was a major challenge, now researchers are faced with an explosion of experimental protocols and bioinformatics tools for producing and analyzing single-cell data.
Scientists at ZS have been working for over a decade to build biological maps that are not only informed by single-cell data but that can also be used to make sense out of single-cell data. Layering cell subtypes, functions, developmental trajectories and spatial organization onto these networks uncovers previously hidden areas of the maps—areas that hold the key for novel therapeutic targets and strategies. Using these maps as a framework over which complex biological data may be laid also enriches single-cell data analysis by providing systems-level context for cellular-level biology. As the single-cell field continues to grow, adding more ‘omic modalities (metabolomics, proteomics and more), these maps will scale to accommodate and extract insight from these new data types.
Effective use of these maps to achieve the end goal—successful therapeutics—also requires combining data across multiple studies in diverse settings to overcome noise and achieve robust, reproducible results. While not easy, this task is achievable with the right mix of biological and technological expertise combined with effective integration with existing data sets.
FIGURE: Progress toward biological maps informed by high-definition, single-cell data
Unblurring the lines between cell types
A diverse team of scientists, including researchers at ZS, Novo Nordisk Foundation Center in Copenhagen, the Broad Institute of MIT and Harvard and the University of Copenhagen combined single-cell technologies and computational analyses to identify a new type of cell, which they call SWAT (structural Wnt-regulated adipose tissue-resident) cells, that shares a progenitor with adipocytes.
Their groundbreaking work, published in Nature Metabolism, leveraged machine-learning-based algorithms to map cells onto existing adipocyte data sets and to order cells in silico by their developmental progress, revealing a clear divergence into two cellular states: an adipogenic branch and a structural branch. These two states possess complementary roles in adipose tissue architecture and function. This work identified specific molecular and genetic signatures associated with each.
By filling in this previously unexplored area of the biological map, researchers have uncovered a hidden player in the regulation of healthy fat tissue. “Leveraging this technology and newly identified player has the potential to transform our understanding of body fat and open new avenues for drug target discovery in cardio-metabolic traits,” says Pascal Timshel, bioinformatics manager at ZS and co-author of the study.
Perhaps the most notable thing about this work is the fact that, although a new cell type was identified, the type of biological samples used in this study have been analyzed for years by the scientific community. This underscores that bulk analyses that average the signal from multiple cell types are a woefully inadequate way to identify biological signal. Single-cell analysis, on the other hand, empowers scientists to analyze their samples at a much higher resolution. Ultimately, this means more-informed drug development efforts with a higher chance of success.
“Now that biopharma R&D has the tools it needs to deeply explore its data—no matter the scale—developing life-saving medicines faster has become a reality.”
Rasmus Wernersson, Principal, ZS Discovery
Introducing the next generation of therapeutic targets
The discovery of new cell types, such as SWAT cells, is an incredible example of the importance of uncovering new areas of the biological map. Doing so lays the groundwork for identifying and leveraging completely new therapeutic paradigms. In the case of SWAT cells, identifying two different cell types originating from a single progenitor opens the door for toggling the balance between these two developmental states to identify new therapeutic targets for cardiometabolic disease—and the concept can be applied to other cell types and other diseases.
Any disease with an immunological component, for example, is characterized by shifts in the proportion of different cell types and demarcations between diseased tissue (where novel disease targets may lie) and invading immune cells. Additionally, neurological disorders are notoriously difficult to diagnose and treat because their molecular mechanisms remain largely a mystery.
We are incredibly excited about the future of drug discovery and development because we believe the solution we need to halt drug failures is already here. As Wernersson said, “Now that biopharma R&D has the tools it needs to deeply explore its data—no matter the scale—developing life-saving medicines faster has become a reality.”
Add insights to your inbox
We’ll send you content you’ll want to read – and put to use.